Computational modeling of virus growth in cells
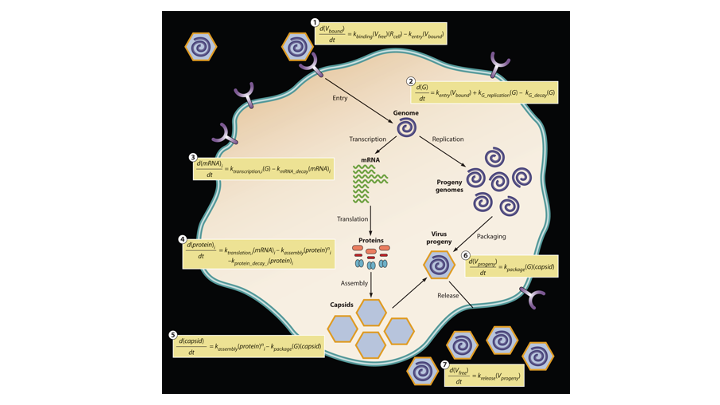
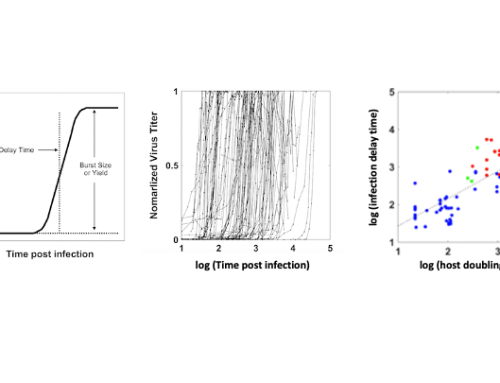
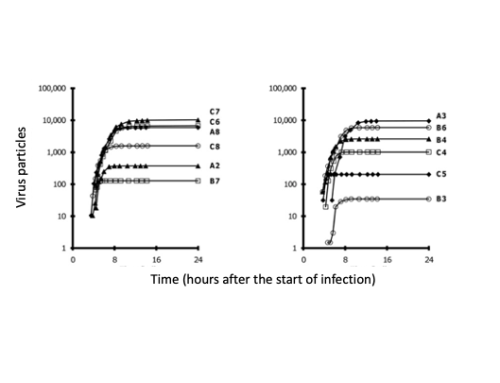
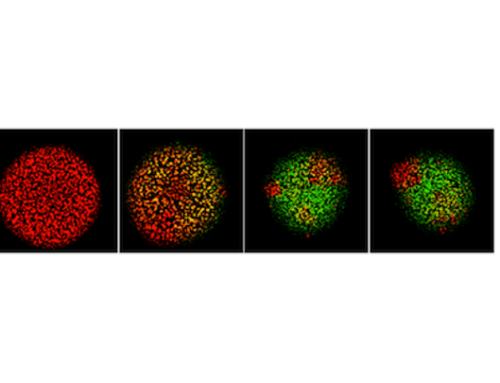
When a virus infects a living cell, how long before virus progeny are released, and how many particles will be made? One may build a model for infection by writing equations that describe each of the essential steps: entry, transcription, translation, genome replication, particle assembly and release from the cell. Experimental data are used to estimate rate parameters for these steps, and computational solution enables one to integrate how the steps interact. We reviewed efforts from over 40 years of such mathematical and computational modeling; see details in Yin and Redovich, 2018.